Abstract
Background: Circulating tumor DNA (ctDNA) has emerged as a promising biomarker for noninvasive disease detection and tumor genotyping in patients with a wide range of malignancies. Circulating tumor DNA often comprises less than 1% of the total cell-free DNA (cfDNA) pool; maximizing recovery of tumor DNA is therefore critical for disease detection and characterization. Levels of ctDNA are dependent upon features of each tumor type, including disease burden (Bettegowda et al., Sci Transl Med 2014; Newman et al., Nat Med 2014). However, ctDNA levels are also potentially sensitive to other analytical considerations, such as the biological source material. In this study, we sought to directly compare plasma and serum samples for ctDNA detection.
Methods: We applied Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) to 33 pretreatment serum and 75 pretreatment plasma samples from patients with diffuse large B cell lymphoma (DLBCL), with matched tumor in 40 cases. In addition, we sequenced matched serum and plasma samples from a single phlebotomy in 3 cases. Germline DNA was also sequenced. Plasma and serum samples were assessed for differences in cfDNA concentration, number of mutations identified, and mean tumor allele fraction (AF). We further compared the size distribution between mutated and wild-type DNA fragments and between plasma and serum samples. Finally, we compared the recovery rate of tumor-biopsy confirmed mutations in plasma and serum samples.
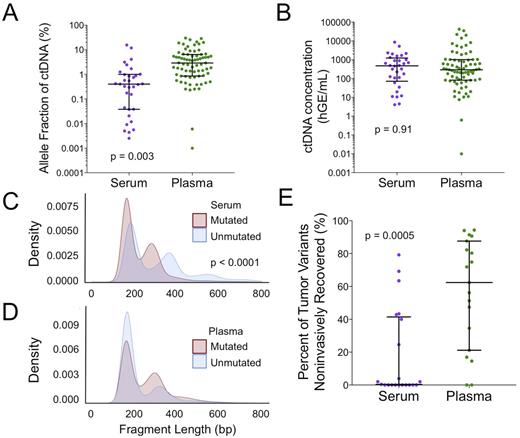
Results: Serum samples had a higher median concentration of cfDNA per mL compared to matched plasma (serum: 481 ng/mL, range 270-499; plasma: 17.7 ng/mL, range 8.4-30). This discrepancy was also seen across the full cohort (serum: 718 ng/mL, IQR 458-1248; plasma: 33.3 ng/mL, IQR 33.3-73.1; p < 0.0001). Despite higher cfDNA concentration, fewer mutations were identified in serum than in plasma in both paired blood samples (serum: 22 mutations per sample, range 0-299; plasma: 186 mutations per sample, range 60-459) and in the full cohort (serum: 50 mutations per sample, IQR 17.8-162; plasma: 184 mutations per sample, IQR 72-331; p < 0.0001). Serum samples also had lower tumor AF than plasma samples in the paired (serum: 0.85 %AF, range 0.71-1.49; plasma 2.8 %AF, range 1.9-8.22) and full cohorts (serum: 0.40 %AF, IQR 0.04-0.98; plasma: 2.91% AF, IQR 0.88-6.05; p = 0.003; Figure 1A). However, when quantitated as a concentration of tumor molecules per mL of sample, serum and plasma had similar levels of ctDNA (serum: 476 haploid genome equivalents (hGE) per mL, IQR 95-1085; plasma: 292 hGE per mL, IQR 82-990; p = 0.91; Figure 1B). This suggests the higher concentration of total cfDNA found in serum consists mostly of non-tumor derived molecules.
In serum, non-mutated molecules were significantly longer compared to molecules harboring tumor mutations (non-mutated: 319 bp, IQR 291-359; mutated: 269 bp, IQR 243-304; p < 0.0001; Figure 1C). This pattern was not seen in plasma (Figure 1D). Interestingly, serum samples also had a longer mean cfDNA fragment length compared to plasma samples (serum: 319 bp, IQR 291-358; plasma: 235 bp, IQR 219-251; p < 0.0001), consistent with the finding that serum contains a higher proportion of non-tumor-derived cfDNA. Finally, when considering mutations identified in tumor biopsies, the percentage of tumor mutations recovered in matched plasma was significantly higher than that in matched serum (serum: 0.4% tumor mutations, IQR 0-40%; plasma: 62% tumor mutations, IQR 28-85%; p = 0.0005; Figure 1E).
Conclusions: Plasma and serum contain equivalent absolute quantities of ctDNA. However, plasma contains a higher fraction of ctDNA than serum, facilitating mutation detection and noninvasive genotyping. When possible, blood samples used for cancer detection should be obtained and stored as plasma to optimize ctDNA recovery.
Figure 1. Comparison of serum and plasma samples.(A) Mean tumor AF is higher in plasma than in serum (p = 0.003) (B) Total quantity of tumor-derived DNA is equivalent between plasma and serum samples (C) In serum, non-tumor-derived cfDNA has a longer average fragment length than ctDNA (p < 0.0001) (D) In plasma, ctDNA and non-tumor-derived cfDNA show overlapping fragment distributions (E) A greater number of mutations seen in tumor biopsies are recovered in paired plasma samples as compared to paired serum samples (p = 0.0005)
Neelapu: Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Bristol-Myers Squibb: Research Funding; Poseida Therapeutics, Inc: Research Funding; Merck: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Karus: Research Funding; Kite Pharma: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Cellectis Inc.: Research Funding. Nastoupil: TG Therapeutics: Honoraria, Research Funding; Genentech: Honoraria, Research Funding; Abbvie: Honoraria, Research Funding; Gilead: Honoraria; Janssen: Honoraria, Research Funding; Karus Therapeutics: Research Funding; Celgene: Honoraria, Research Funding. Diehn: Novartis: Consultancy; Roche: Consultancy; Quanticel Pharmaceuticals: Consultancy; Varian Medical Systems: Research Funding. Alizadeh: Roche: Consultancy; Genentech: Consultancy; Celgene: Consultancy; CiberMed: Consultancy; Gilead: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal